Martin Sauzade PhD.
Resume
 Single-cell technologies are extending our comprehension of biological systems by bringing the study of biological phenomena to the cellular level. For the purpose of cellular distinction, most technologies require the isolation of individual cells. This compartmentalization is most conveniently accomplished by encapsulating single-cells within micro droplets using a microfluidic device. In the last three years, technologies based on the high throughput generation of droplets have enabled the analysis of samples with more than 100k cells. However, for numerous clinical samples such as biopsies or circulating tumor cells, it can be difficult to obtain such a sample size.
Single-cell technologies are extending our comprehension of biological systems by bringing the study of biological phenomena to the cellular level. For the purpose of cellular distinction, most technologies require the isolation of individual cells. This compartmentalization is most conveniently accomplished by encapsulating single-cells within micro droplets using a microfluidic device. In the last three years, technologies based on the high throughput generation of droplets have enabled the analysis of samples with more than 100k cells. However, for numerous clinical samples such as biopsies or circulating tumor cells, it can be difficult to obtain such a sample size.
I addressed this shortcoming during my tenure as a postdoctoral researcher at Stony Brook University, where I used innovative strategies to conduct single-cell encapsulation within a microfluidic device. I designed and developed multiple approaches to conduct the encapsulation of individual cells in a low throughput, highly controllable manner while satisfying the constraints inherent to a downstream ‘omic’ analysis.
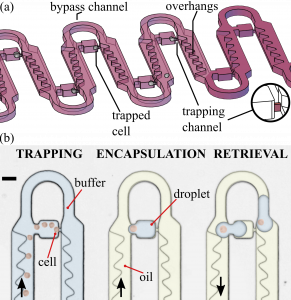
One of the platforms that I developed was recently presented in a Lab on a Chip communication entitled “Deterministic trapping, encapsulation and retrieval of single-cells”. This microfluidic chip relies on the trapping of single cells prior to their encapsulation through an interplay between hydrodynamics and wetting dynamics. The device efficiently encapsulates hundreds of individual cells into droplets while exhibiting no sample loss, effectively laying the foundations for the single-cell analysis of rare samples via droplet microfluidics.