Predicted Transcription Factor Binding Sites:
To develop broadly useful methods for the genetic manipulation of Labyrinthulomycetes, it is essential to understand the similarities and differences in regulation of gene expression among them. Toward this end we have used FIMO from the MEME suite (http://meme-suite.org/doc/fimo.html) to identify motifs similar to yeast transcription factor binding sites in each of the three available genome sequences: Aplanochytrium kerguelense PBS07, Schizochytrium aggregatum ATCC 28209, and Aurantiochytrium limacinum ATCC MYA-1381.
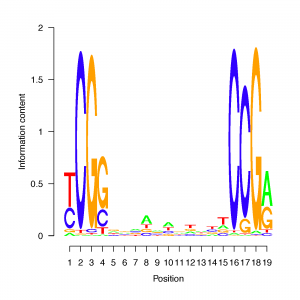
The logos below illustrate yeast-like GAL4 binding sites in each of the three genomes. The main six positions of the motif (CGG-N11-CCG) are conserved, while the intermediate positions reflect the overall GC content of each (lowest for Aplanochytrium, intermediate for Aurantiochytrium, and highest for Schizochytrium).
The scripts used for motif discovery in this analysis are published at protocols.io, and the raw results of the motif search can be found at Academic Commons.

Kozak Sequences:
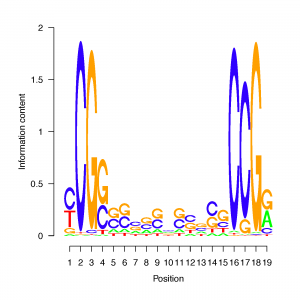
We are comparing the 50 bp sequences surrounding predicted translation initiation sites in the three available labyrinthulomycete genome sequences: Aplanochytrium kerguelense PBS07, Schizochytrium aggregatum ATCC 28209, and Aurantiochytrium limacinum ATCC MYA-1381. The figure below shows logos of the most common motifs in each genome. There are distinct differences among strains that are related to the difference in GC content noted for their CDS regions (see Codon Usage page).
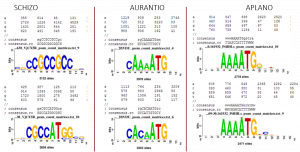
The most common motifs in Aplanochytrium and Aurantiochytrium resemble previously described fungal or protistan Kozak sequences. The most common motifs from Schizochytrium are distinctly different, with the bottom one more similar to a canonical vertebrate Kozak sequence. The most common Schizochytrium motif does not include the ATG, instead being found as potentially hairpin-forming sequences on either side.The scripts used for evaluation of Kozak sequence over-representation are published at protocols.io.
We are currently assessing the statistical significance of secondary structures predicted around the ATG for Schizochytrium. The scripts used for evaluating secondary structure significance can be be found at protocols.io.