MAR581: Applications of Next Generation Sequencing in Functional Genomics
Course Description: Integrative genomics is a new area of research that seeks to understand the structure and function of an organism’s genes. This course provides an integrated view of how these methods can be used to answer questions regarding marine organisms evolution, biology and ecology. Over the course of the semester, examples will cover various topics including marine biodiversity, population structures, environmental adaptation, stress responses, phylogeny of animals, Aquaculture and fisheries, interaction between species (predation, parasitism, mutualism…). A particular accent is given to the role of Next Generation Sequencing technologies in answering questions related to these topics.
This course prepares the new generation of students to the use of new technologies, regardless of the research topic they are interested in. At the interface between ecology, evolution, cellular biology, microbiology and genomics, the course of functional and integrative marine genomics addresses Stony Brook University goal of an interdisciplinary graduate education. The course will familiarize students with the latest techniques and prepare them to adapt to the ever-evolving technologies as they conduct their own research. This course is an attractive addition to the curriculum within the graduate program for all students interested in biological sciences.
LEARNING OBJECTIVES:
By the end of the semester students shall be able to
- Define terms used in structural genomics and functional genomics
- Understand how genomics methods work
- Interpret data presented in a scientific paper and evaluate its content
- Read and understand the operating manuals of NGS tools
- Choose between different methods to answer a biological question
- Acquire a broad knowledge of how these methods can be used to answer questions related to the evolution, ecology and biology of marine organisms
These objectives are met through participation in lectures, reading assignments, discussions, practical bioinformatics sessions, preparing and presenting a literature review, and preparing illustrated fact sheets on various topics. When possible, the course also includes a visit to a Genome Center and discussion with some of its members.
Topics covered (in order over the course of the semester): Second and Third generation sequencing techniques, Genome assembly, Comparative genomics, Population genomics, Epigenomics, Transcriptomics, Metagenomics and Metatranscriptomics
Fact Sheets from Fall 2016 students
 Detecting epigenetic modifications by single molecule real time (SMRT) sequencing
Detecting epigenetic modifications by single molecule real time (SMRT) sequencing
Genomics shed light on Mysteries of evolution
 Methods to fragment DNA for Next generation sequencing
Methods to fragment DNA for Next generation sequencing
 RADseq A powerful new tool for Next generation applications
RADseq A powerful new tool for Next generation applications
 Whole genome resequencing reveals population history
Whole genome resequencing reveals population history
 Connecting Epigenetics and Phenotypic Variation in Ecology
Connecting Epigenetics and Phenotypic Variation in Ecology
 Applications of transcriptomics in Ecotoxicology
Applications of transcriptomics in Ecotoxicology
 The Marine Eukaryote Transcriptome Sequencing Project is an incredible1
The Marine Eukaryote Transcriptome Sequencing Project is an incredible1
 Sumoylation and repression of gene expression
Sumoylation and repression of gene expression
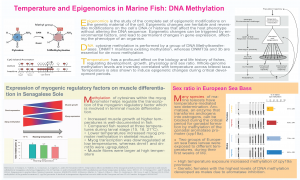 Temperature and epigenomics in marine fish
Temperature and epigenomics in marine fish
 Exploring the three-dimensioinal organization of genomes
Exploring the three-dimensioinal organization of genomes
Fact Sheets from Fall 2015 students
 Multivariate Statistics in Marine Genomics
Multivariate Statistics in Marine Genomics
 What’s eating you? Using Next Generation Sequencing in Forensic Science
What’s eating you? Using Next Generation Sequencing in Forensic Science
 Microbiome importance in the plant holobiont
Microbiome importance in the plant holobiont
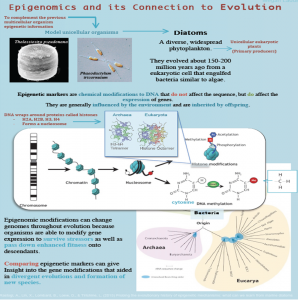 Epigenomics and its connection to Evolution
Epigenomics and its connection to Evolution
 Visualizing Wholed Genome Chromosome Structure
Visualizing Wholed Genome Chromosome Structure
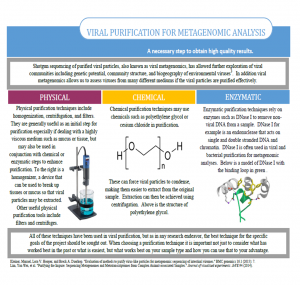 Viral purification for metagenomic analysis
Viral purification for metagenomic analysis
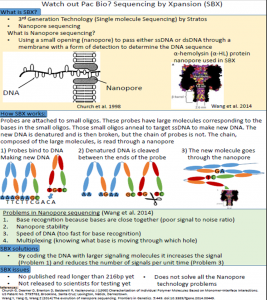 Watch out Pac Bio? Sequencing by Xpansion (SBX)
Watch out Pac Bio? Sequencing by Xpansion (SBX)
 mRNA enrichment techniques for Prokaryotes
mRNA enrichment techniques for Prokaryotes













CompTIA Security+ SY0-701 Dumps Cafe is a valuable certification that requires diligent preparation and adherence to ethical standards. While dumps cafes may offer tempting shortcuts, the risks associated with using exam dumps far outweigh any potential benefits. Candidates should prioritize ethical certification practices and utilize legitimate study resources to ensure their success in the Security+ SY0-701 exam and beyond.